| CROSS-DISCIPLINARY PHYSICS AND RELATED AREAS OF SCIENCE AND TECHNOLOGY |
 |
|
|
|
|
Accurate Evaluation on the Interactions of SARS-CoV-2 with Its Receptor ACE2 and Antibodies CR3022/CB6 |
| Hong-ming Ding1*, Yue-wen Yin1, Song-di Ni1, Yan-jing Sheng1, and Yu-qiang Ma2* |
1Center for Soft Condensed Matter Physics and Interdisciplinary Research, School of Physical Science and Technology, Soochow University, Suzhou 215006, China
2National Laboratory of Solid State Microstructures and Department of Physics, Collaborative Innovation Center of Advanced Microstructures, Nanjing University, Nanjing 210093, China
|
|
| Cite this article: |
|
Hong-ming Ding, Yue-wen Yin, Song-di Ni et al 2021 Chin. Phys. Lett. 38 018701 |
|
|
|
|
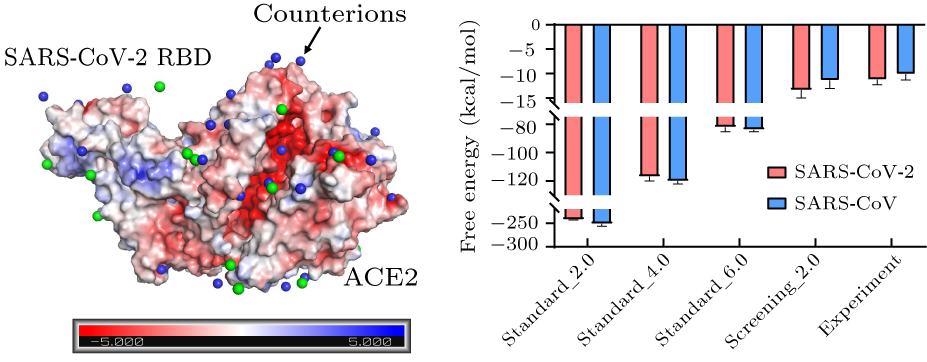
Abstract The spread of the coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) has become a global health crisis. The binding affinity of SARS-CoV-2 (in particular the receptor binding domain, RBD) to its receptor angiotensin converting enzyme 2 (ACE2) and the antibodies is of great importance in understanding the infectivity of COVID-19 and evaluating the candidate therapeutic for COVID-19. We propose a new method based on molecular mechanics/Poisson–Boltzmann surface area (MM/PBSA) to accurately calculate the free energy of SARS-CoV-2 RBD binding to ACE2 and antibodies. The calculated binding free energy of SARS-CoV-2 RBD to ACE2 is $-13.3$ kcal/mol, and that of SARS-CoV RBD to ACE2 is $-11.4$ kcal/mol, which agree well with the experimental results of $-11.3$ kcal/mol and $-10.1$ kcal/mol, respectively. Moreover, we take two recently reported antibodies as examples, and calculate the free energy of antibodies binding to SARS-CoV-2 RBD, which is also consistent with the experimental findings. Further, within the framework of the modified MM/PBSA, we determine the key residues and the main driving forces for the SARS-CoV-2 RBD/CB6 interaction by the computational alanine scanning method. The present study offers a computationally efficient and numerically reliable method to evaluate the free energy of SARS-CoV-2 binding to other proteins, which may stimulate the development of the therapeutics against the COVID-19 disease in real applications.
|
|
Received: 06 November 2020
Published: 18 December 2020
|
|
|
|
|
| Fund: Supported by the National Natural Science Foundation of China (Grant Nos. 11874045 and 11774147). |
|
|
|
| [1] | https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/ |
| [2] | Zhou P, Yang X L, Wang X G, Hu B, Zhang L, Zhang W, Si H R, Zhu Y, Li B, Huang C L, Chen H D, Chen J, Luo Y, Guo H, Jiang R D, Liu M Q, Chen Y, Shen X R, Wang X, Zheng X S, Zhao K, Chen Q J, Deng F, Liu L L, Yan B, Zhan F X, Wang Y Y, Xiao G F and Shi Z L 2020 Nature 579 270 |
| [3] | Wu F, Zhao S, Yu B, Chen Y M, Wang W, Song Z G, Hu Y, Tao Z W, Tian J H, Pei Y Y, Yuan M L, Zhang Y L, Dai F H, Liu Y, Wang Q M, Zheng J J, Xu L, Holmes E C and Zhang Y Z 2020 Nature 579 265 |
| [4] | Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao G F and Tan W 2020 New Engl. J. Med. 382 727 |
| [5] | Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu W J, Wang D, Xu W, Holmes E C, Gao G F, Wu G, Chen W, Shi W and Tan W 2020 Lancet 395 565 |
| [6] | Walls A C, Park Y J, Tortorici M A, Wall A, McGuire A T and Veesler D 2020 Cell 181 281 |
| [7] | Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens T S, Herrler G, Wu N H, Nitsche A, Müller M A, Drosten C and Pöhlmann S 2020 Cell 181 271 |
| [8] | Cai Y F, Zhang J, Xiao T S, Peng H Q, Sterling S M, Walsh R M, Rawson S, Rits-Volloch S and Chen B 2020 Science 369 1586 |
| [9] | Xia S, Zhu Y, Liu M, Lan Q, Xu W, Wu Y, Ying T, Liu S, Shi Z, Jiang S and Lu L 2020 Cell. & Mol. Immunol. 17 765 |
| [10] | Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L and Wang X 2020 Nature 581 215 |
| [11] | Shi R, Shan C, Duan X, Chen Z, Liu P, Song J, Song T, Bi X, Han C, Wu L, Gao G, Hu X, Zhang Y, Tong Z, Huang W, Liu W J, Wu G, Zhang B, Wang L, Qi J, Feng H, Wang F S, Wang Q, Gao G F, Yuan Z and Yan J 2020 Nature 584 120 |
| [12] | Wang C, Li W, Drabek D, Okba N M A, van Haperen R, Osterhaus A D M E, van Kuppeveld F J M, Haagmans B L, Grosveld F and Bosch B J 2020 Nat. Commun. 11 2251 |
| [13] | Yuan M, Wu N C, Zhu X, Lee C C D, So R T Y, Lv H, Mok C K P and Wilson I A 2020 Science 368 630 |
| [14] | Long Q X, Liu B Z, Deng H J, Wu G C, Deng K, Chen Y K, Liao P, Qiu J F, Lin Y, Cai X F, Wang D Q, Hu Y, Ren J H, Tang N, Xu Y Y, Yu L H, Mo Z, Gong F, Zhang X L, Tian W G, Hu L, Zhang X X, Xiang J L, Du H X, Liu H W, Lang C H, Luo X H, Wu S B, Cui X P, Zhou Z, Zhu M M, Wang J, Xue C J, Li X F, Wang L, Li Z J, Wang K, Niu C C, Yang Q J, Tang X J, Zhang Y, Liu X M, Li J J, Zhang D C, Zhang F, Liu P, Yuan J, Li Q, Hu J L, Chen J and Huang A L 2020 Nat. Med. 26 845 |
| [15] | Wrapp D, De Vlieger D, Corbett K S, Torres G M, Wang N, Van Breedam W, Roose K, van Schie L, Hoffmann M, Pöhlmann S, Graham B S, Callewaert N, Schepens B, Saelens X and McLellan J S 2020 Cell 181 1004 |
| [16] | Yuan M, Liu H, Wu N C, Lee C C D, Zhu X, Zhao F, Huang D, Yu W, Hua Y, Tien H, Rogers T F, Landais E, Sok D, Jardine J G, Burton D R and Wilson I 2020 Science 369 1119 |
| [17] | Poh C M, Carissimo G, Wang B, Amrun S N, Lee C Y P, Chee R S L, Fong S W, Yeo N K W, Lee W H, Torres-Ruesta A, Leo Y S, Chen M I C, Tan S Y, Chai L Y A, Kalimuddin S, Kheng S S G, Thien S Y, Young B E, Lye D C, Hanson B J, Wang C I, Renia L and Ng L F P 2020 Nat. Commun. 11 2806 |
| [18] | Wang Y, Liu M and Gao J 2020 Proc. Natl. Acad. Sci. USA 117 13967 |
| [19] | Spinello A, Saltalamacchia A and Magistrato A 2020 J. Phys. Chem. Lett. 11 4785 |
| [20] | Amin M, Sorour M K and Kasry A 2020 J. Phys. Chem. Lett. 11 4897 |
| [21] | Othman H, Bouslama Z, Brandenburg J T, da R J, Hamdi Y, Ghedira K, Srairi-Abid N and Hazelhurst S 2020 Biochem. Biophys. Res. Commun. 527 702 |
| [22] | Han Y and Král P 2020 ACS Nano 14 5143 |
| [23] | Qiao B and de Olvera L C M 2020 ACS Nano 14 10616 |
| [24] | Xu X, Chen P, Wang J, Feng J, Zhou H, Li X, Zhong W and Hao P 2020 Sci. Chin. Life Sci. 63 457 |
| [25] | Wang J, Xu X, Zhou X, Chen P, Liang H, Li X, Zhong W and Hao P 2020 J. Gen. Virol. 101 921 |
| [26] | Zou J, Yin J, Fang L, Yang M, Wang T, Wu W, Bellucci M A and Zhang P 2020 J. Chem. Inf. Model. |
| [27] | Sun H, Li Y, Tian S, Xu L and Hou T 2014 Phys. Chem. Chem. Phys. 16 16719 |
| [28] | Chen F, Sun H, Wang J, Zhu F, Liu H, Wang Z, Lei T, Li Y and Hou T 2018 RNA 24 1183 |
| [29] | Chodera J D, Mobley D L, Shirts M R, Dixon R W, Branson K and Pande V S 2011 Curr. Opin. Struct. Biol. 21 150 |
| [30] | Lin Y L, Aleksandrov A, Simonson T and Roux B 2014 J. Chem. Theory Comput. 10 2690 |
| [31] | Rocklin G J, Mobley D L, Dill K A and Hünenberger P H 2013 J. Chem. Phys. 139 184103 |
| [32] | Wrapp D, Wang N, Corbett K S, Goldsmith J A, Hsieh C L, Abiona O, Graham B S and McLellan J S 2020 Science 367 1260 |
| [33] | Li F, Li W, Farzan M and Harrison S C 2005 Science 309 1864 |
| [34] | Jorgensen W L and Madura J D 1983 J. Am. Chem. Soc. 105 1407 |
| [35] | Basdevant N, Weinstein H and Ceruso M 2006 J. Am. Chem. Soc. 128 12766 |
| [36] | Venken T, Krnavek D, Münch J, Kirchhoff F, Henklein P, De Maeyer M and Voet A 2011 Proteins: Struct. Funct. Bioinf. 79 3221 |
| [37] | Abraham M J, Murtola T, Schulz R, Páll S, Smith J C, Hess B and Lindahl E 2015 SoftwareX 1–2 19 |
| [38] | Maier J A, Martinez C, Kasavajhala K, Wickstrom L, Hauser K E and Simmerling C 2015 J. Chem. Theory Comput. 11 3696 |
| [39] | Hess B, Bekker H, Berendsen H J C and Fraaije J G E M 1997 J. Comput. Chem. 18 1463 |
| [40] | Essmann U, Perera L, Berkowitz M L, Darden T, Lee H and Pedersen L G 1995 J. Chem. Phys. 103 8577 |
| [41] | Genheden S and Ryde U 2015 Expert. Opin. Drug. Dis. 10 449 |
| [42] | Wang E, Sun H, Wang J, Wang Z, Liu H, Zhang J Z H and Hou T 2019 Chem. Rev. 119 9478 |
| [43] | Debye P and Hückel E 1923 Physikal. Z. 24 185 |
| [44] | Levin Y 2002 Rep. Prog. Phys. 65 1577 |
| [45] | Sun Z, Yan Y N, Yang M and Zhang J Z H 2017 J. Chem. Phys. 146 124124 |
| [46] | Duan L, Liu X and Zhang J Z H 2016 J. Am. Chem. Soc. 138 5722 |
| [47] | https://jerkwin.github.io/gmxtool |
| [48] | Massova I and Kollman P A 1999 J. Am. Chem. Soc. 121 8133 |
| [49] | Moreira I S, Fernandes P A and Ramos M J 2007 J. Comput. Chem. 28 644 |
| [50] | Huo S G, Massova I and Kollman P A 2002 J. Comput. Chem. 23 15 |
| [51] | Harapan H, Itoh N, Yufika A, Winardi W, Keam S, Te H, Megawati D, Hayati Z, Wagner A L, Mudatsir M 2020 J. Infect. Public Health 13 667 |
| [52] | Liu Y, Gayle A A, Wilder-Smith A and Rocklöv J 2020 J. Travel Med. 27 taaa021 |
| [53] | Kepler T B and Perelson A S 1993 J. Theor. Biol. 164 37 |
|
|
Viewed |
|
|
|
Full text
|
|
|
|
|
Abstract
|
|
|
|
|